Nanosystems: Phys. Chem. Math., 2022, 13 (3), 274–284
Assessment of structural changes in proteins and surrounding water molecules in solution according to SAXS and MD data
Alexander V. Smirnov – ITMO University, Saint Petersburg 197101, Russia; asmirnav_2@mail.ru
Andrei M. Semenov – Alferov University Russian Academy of Sciences, Saint Petersburg 194021; Voronezh State University, Voronezh 394018, Russia
Yuri B. Porozov – Sechenov First Moscow State Medical University, Moscow 119991; Sirius University of Science and Technology, Sochi 354340, Russia
Boris A. Fedorov – ITMO University, Saint Petersburg 197101, Russia
Corresponding author: A.V. Smirnov, smirnav_2@mail.ru
DOI 10.17586/2220-8054-2022-13-3-274-284
PACS 61.05.cf, 71.15. Pd, 87.15.B
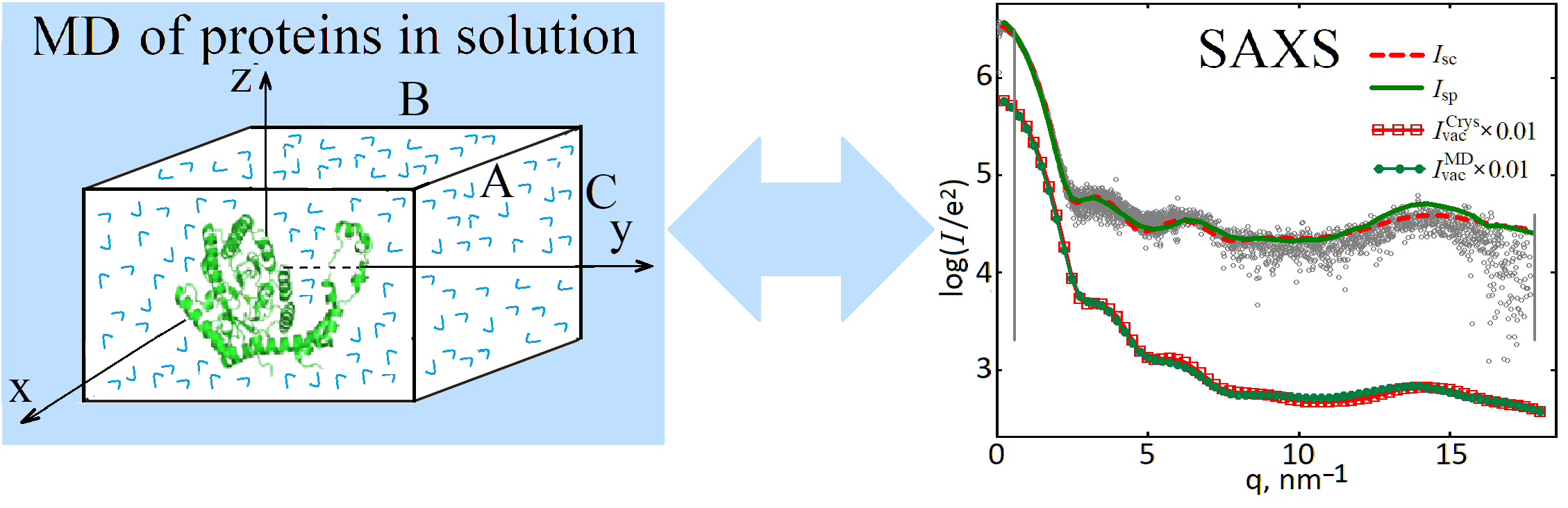
ABSTRACT The SASPAR program for calculation of SAXS of proteins in solution uses trajectories of molecular dynamics (MD) and an explicit solvent model. The program allows one to take into account real interactions of solvent molecules both between each other and with the protein molecule. The previously developed SASCUBE program (the “cube method”) is also used, it assumes that the protein structures in crystal and in solution coincide, and the water surrounding the proteins is considered as a homogeneous continuum. Using these programs, SAXS curves were calculated for 18 proteins of different molecular weights and then compared with one another and with the corresponding experimental scattering curves. “Vacuum” SAXS curves (i.e., without taking into account the surrounding water) were also calculated for each protein for two approaches: a) based on the coordinates of protein atoms in crystal and b) based on the coordinates of protein atoms for each MD frame with further averaging of the intensities from all the frames. 1) It was shown that for the 14 single-domain proteins considered, the “vacuum” scattering curves calculated by two methods coincide well for almost each protein. Hence, the structure of the studied proteins in a solution is similar to their structure in a crystal and, therefore, the presence of the surrounding water molecules does not alter the protein structure itself significantly. The SASPAR- and SASCUBE-curves coincide well only in two cases (i.e., water is only slightly structured near the protein surface), but in the other cases these curves are markedly different, which indicates the structuredness of the water near the protein surface, although to a different extent. 2) It was shown that for the 4 multi-domain proteins considered, their “vacuum” scattering curves, calculated with the two methods indicated above, differ noticeably, which is an evidence that their crystalline and “water” structures are different. It was also shown that the most of the calculated curves coincide well with the experimental
ones.
KEYWORDS small-angle X-ray solution scattering, molecular dynamics, protein structure in solution, water structure
ACKNOWLEDGEMENTS The authors are grateful to Dr. Alexander Grishaev for providing the experimental SAXS scattering curves of the GB3 protein and to the Master’s student K.S. Mazokha for the assistance with research.
FOR CITATION Smirnov A.V., Semenov A.M., Porozov Yu.B., Fedorov B.A. Assessment of structural changes in proteins and surrounding water molecules in solution according to SAXS and MD data. Nanosystems: Phys. Chem. Math., 2022, 13 (3), 274–284.
[In Russian] А.В. Смирнов, А.М. Семенов, Ю.Б. Порозов, Б.А. Федоров
Оценка структурных изменений в белках и окружающих молекул воды в растворе по данным SAXS и MD
АННОТАЦИЯ Программа SASPAR для расчета МУРР белков в растворе использует траектории молекулярной динамики (МД) и явную модель растворителя. Программа позволяет учитывать реальные взаимодействия молекул растворителя как между собой, так и с молекулой белка. Также используется разработанная ранее программа SASCUBE («метод куба»), в ней предполагается, что структуры белков в кристалле и в растворе совпадают, а вода, окружающая белки, рассматривается как однородный континуум. С помощью этих программ были рассчитаны кривые МУРР для 18 белков различной молекулярной массы, а затем сопоставлены друг с другом и с соответствующими экспериментальными кривыми рассеяния. «Вакуумные» кривые МУРР (т. е. без учета окружающей воды) также рассчитывались для каждого белка по двум подходам: а) по координатам атомов белка в кристалле и б) по координатам атомов белка для каждой МН. кадр с последующим усреднением интенсивностей со всех кадров. 1) Показано, что для 14 рассмотренных однодоменных белков кривые «вакуумного» рассеяния, рассчитанные двумя методами, хорошо совпадают практически для каждого белка. Следовательно, структура исследованных белков в растворе аналогична их структуре в кристалле, и, следовательно, присутствие окружающих молекул воды не изменяет существенно саму структуру белка. SASPAR- и SASCUBE-кривые хорошо совпадают только в двух случаях (т. е. вода слабо структурирована вблизи поверхности белка), но в остальных случаях эти кривые заметно различаются, что свидетельствует о структурированности воды вблизи поверхности белка, т.е. хотя и в разной степени. 2) Показано, что для 4-х рассмотренных многодоменных белков их «вакуумные» кривые рассеяния, рассчитанные двумя указанными выше методами, заметно различаются, что свидетельствует о различии их кристаллической и «водной» структур. Также было показано, что большинство расчетных кривых хорошо совпадают с экспериментальными.
КЛЮЧЕВЫЕ СЛОВА малоугловое рассеяние рентгеновских лучей в растворе, молекулярная динамика, структура белка в растворе, структура воды.