NANOSYSTEMS: PHYSICS, CHEMISTRY, MATHEMATICS, 2019, 10 (6), P. 701–710
The effect of carbon nanotube on the structure of H–NS protein DNA complex: molecular dynamics approach
Najmeh Mahdavipour – Department of Chemistry, Mashhad Branch, Islamic Azad University, Mashhad, Iran
Mohammad Reza Bozorgmehr – Department of Chemistry, Mashhad Branch, Islamic Azad University, Mashhad, Iran; bozorgmehr@mshdiau.ac.ir; mr_bozorgmehr@yahoo.com
Mohammad Momen-Heravi – Department of Chemistry, Mashhad Branch, Islamic Azad University, Mashhad, Iran
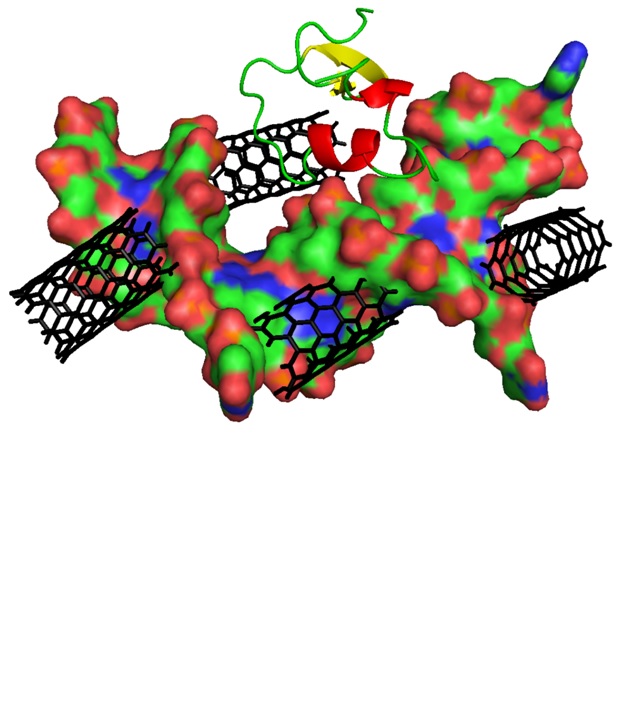
Most of the experimental biophysical and biochemical observations of proteins are in dilute solutions, while inside the cell is a crowded environment. The effect of crowding on the structure and activity of biomolecules is not completely clear. In this work, molecular dynamics simulation was used to study the effect of single walled carbon nanotube (SWCNT) on the H–NS protein in the presence and absence of double-stranded nucleic acid. The values of root mean square deviation (RMSD) and its distribution, radius of gyration (Rg) and its distribution and root mean square fluctuation (RMSF) were calculated. Changes in the secondary structure of the H–NS were also calculated. The contributions of each residue of H–NS in free energy of binding between H–NS and DNA were calculated. The results indicate that the SWCNT unfolds the structure of the H–NS. In terms of contribution of residues in secondary structures, in the presence of a SWCNT, the sheet secondary structure of the H–NS changes more than helices secondary structure. In the triple system, which includes H–NS, SWCNT and DNA; Ala-1, Arg-3, Lys-6, Lys-17, Arg-24, Lys-30, Lys-31, Lys-38 and Lys-46 residues have a favorable effect on the interaction of the H–NS with the DNA.
Keywords: crowding, MMPBSA, secondary structure, contact map.
DOI 10.17586/2220-8054-2019-10-6-701-710